How to Use Docker to Set Up a Jupyter Server for Data Science
Learning data science is exciting, but setting up the right environment can quickly become a frustrating challenge. At Dataquest, we provide an easy-to-use, preconfigured environment with Python, popular data science libraries, and an interactive code editor. This makes it simple for beginners and experienced data scientists to start coding immediately. However, when it’s time to work with your own datasets, transitioning to a local setup—or better yet, running a Dockerized environment for data science—is often the next step.
Unfortunately, setting up a local environment can be one of the most discouraging parts of the process. Inconsistent package versions, installation errors, and platform-specific setup issues can leave you stuck before you even begin. These problems are magnified when collaborating with teams using different operating systems. That’s why using Docker for data science is a game-changer—it provides a consistent, portable data science setup that eliminates the hassle. In this post, we’ll show you how to run a Docker environment with Jupyter and get back to what matters: solving data problems.
Fortunately, there has been a rise of technologies that help with these development woes. The one we'll be exploring in this post is a containerization tool called Docker. Since 2013, it has made it fast and easy to launch multiple Docker data science environments supporting the infrastructure needs of different projects.
In this tutorial, we're going to show you how to set up your own Jupyter Notebook server using Docker to create your own customized data science environment. We'll cover the basics of Docker and containerization, how to install Docker, and how to download and run Dockerized applications. By the end, you should be able to run your own local Jupyter server with the latest data science libraries.
The Docker whale is here to help!

An Overview of Docker and Containerization
Before we dive into Docker, it's important to know some preliminary software concepts that led to the rise of technologies like Docker. In the introduction, we briefly described the difficulty of working on teams with multiple operating systems and installing third-party libraries. These types of problems have been around since the beginning of software development.
One solution has been the use of virtual machines. Virtual machines allow you to emulate alternative operating systems from the one running on your local machine. A common example is running a Windows desktop with a Linux virtual machine. A virtual machine is essentially a fully isolated operating system with applications that are run independent of your own system. This is extremely helpful in a team of developers as every member can run the exact same system regardless of the OS on their machine.
However, virtual machines are not a panacea. They are difficult to set up, require significant system resources to run, and take a long time to boot.

An example of using Windows in a virtual machine on a mac
Building on this concept of virtualization, an alternative approach to full virtual machine isolation is called containerization. Containers are similar to virtual machines as they also run applications in an isolated environment. However, instead of running a complete operating system with all of its libraries, a containerized environment is a lightweight process that runs on top of a container engine.
The container engine runs the containers and provides the ability for the host OS to share read only libraries and its kernel to the container. This makes a running container MBs in size versus virtual machines that can be tens of GBs or more. Docker is a type of container engine. It allows us to download and run images which contain a preconfigured lightweight OS, set of libraries, and application specific packages. When we run an image, the process spawned by the Docker engine is called a container.
As mentioned earlier, containers eliminate configuration problems and ensure compatibility across platforms, freeing us from the restrictions of underlying operating systems or hardware. Similar to virtual machines, systems built on different technologies (e.g. Mac OS X vs. Microsoft Windows) can deploy completely identical containers. There are multiple advantages to using containers over virtual environments:
- Ability to get started quickly. You don’t need to wait for packages to install when you just want to jump in and start doing analysis.
- Consistent across platforms. Python packages are cross-platform, but some behave differently on Windows vs Linux, and some have dependencies that can’t be installed on Windows. Docker containers always run in a Linux environment, so they’re consistent.
- Ability to checkpoint and restore. You can install packages into a Docker image, then create a new image of that checkpoint. This gives you the ability to quickly undo changes or rollback configurations.
A good overview of containerization and the difference between virtual machines can be found in the official Docker documentation. In the next section, we're going to cover how to setup and run Docker on your system.
Installing Docker
There’s a graphical installer for Windows and Mac that makes installing Docker easy. Here are instructions for each OS:
For the rest of the tutorial we'll be covering the Linux and macOS (Unix) instructions of running Docker to create a data science environment. The examples we'll provide should be run in your terminal application (Unix), or the DOS prompt (Windows). While we'll highlight the Unix shell commands, the Windows commands should be similar. We recommend checking the official Docker documentation if there is any discrepancy. To check that the Docker client is correctly installed, here are a few test commands:
docker version: Returns information on the Docker version running on your local machine.docker help: Returns a list of Docker commands.docker run hello-world: Runs the hello-world image and verifies that Docker is correctly installed and functioning.
Running a Docker Container from an Image
With Docker installed, we can now download and run images. Recall that an image contains a lightweight OS and libraries to run your application, while the running image is called a container. You can think of an image as the executable file, and the running process spawned by that file as the container. Let's start by running a basic Docker image.
Enter the docker run command (below) in your shell prompt. Make sure to enter the full command:
docker run ubuntu:16.04If your Docker is installed correctly, you should see something like the following output:
Unable to find image 'ubuntu:16.04' locally
16.04: Pulling from library/ubuntu
297061f60c36: Downloading [============> ] 10.55MB/43.03MB
e9ccef17b516: Download complete
dbc33716854d: Download complete
8fe36b178d25: Download complete 686596545a94: Download completeLet's break down the previous command. First, we started by passing the run argument to the docker engine. This tells Docker that the next argument, ubuntu:16.04 is the image we want to run. The image argument we passed in is composed of the image name, ubuntu and a corresponding tag, 16.04. You can think of the tag as the image version. Furthermore, if you were to leave the image tag blank, Docker would run the latest image version (i.e. docker run ubuntu < -> docker run ubuntu:latest).
Once we issue the command, Docker starts the run process by checking if the image is on your local machine. If Docker can't find the image, it will check Docker Hub and download the image. Docker hub is an image repository, meaning it hosts open source community built images that are available to download.
Finally, after downloading the image, Docker will then run it as a container. However, notice that when the ubuntu container starts up, it immediately exits. The reason it exits is because we didn't pass in additional arguments providing context to the running container. Let's try running another image with some optional arguments to the run command. In the following command, we'll provide the -i and -t flag, starting an interactive session and simulated terminal (TTY).
Run the following to get access to a Python prompt running in a Docker container:
docker run -i -t python:3.6This is equivalent to:
docker run -it python:3.6Running the Jupyter Notebook
After running the previous command, you should have entered the Python prompt. Within the prompt, you can write Python code as normal but the code will be exectuting in the running Docker container. When you exit the prompt, you'll both quit the Python process and leave the interactive container mode which shuts down the Docker container. So far, we have run both an Ubuntu and Python image. These types of images are great to develop with, but they're not that exciting on their own. Instead, we're going to run a Jupyter image that is an application specific image that is built on top of the ubuntu image.
The Jupyter images we'll be using come from Jupyter's development community. The blueprint of the images, called a Dockerfile, can be found in their Github repo. We won't cover Dockerfiles in detail this tutorial, so just think of them as the source code for the created image. An image's Dockerfile is commonly hosted on Github while the built image is hosted on Docker Hub. To begin, let's call the Docker run command on one of the Jupyter images.
We're going to run the minimal-notebook that only has Python and Jupyter installed. Enter the command below: docker run jupyter/minimal-notebook Using this command, we'll be pulling the latest image of the minimal-notebook from the jupyter Docker hub account. You'll know it has ran successfully if you see the following output:
[C 06:14:15.384 NotebookApp]
Copy/paste this URL into your browser when you connect for the first time,
to login with a token:
https://localhost:8888/?token=166aead826e247ff182296400d370bd08b1308a5da5f9f87Similar to running a non-Dockerized Jupyter notebook, you'll have a link to the Jupyter localhost server and a given token. However, if you try to navigate to the provided link, you won't be able to access the server. On your browser, you'll be presented instead with a "Site Cannot be Reached" page.
This is because the Jupyter server is running within it's own isolated Docker container. This means that all the ports, directories, or any other files are not shared with your local machine unless explicitly directed. To access the Jupyter server in the Docker container, we need to open the ports between the host and container by passing in the -p <host_port>:<container_port> flag and argument.
docker run -p 8888:8888 jupyter/minimal-notebook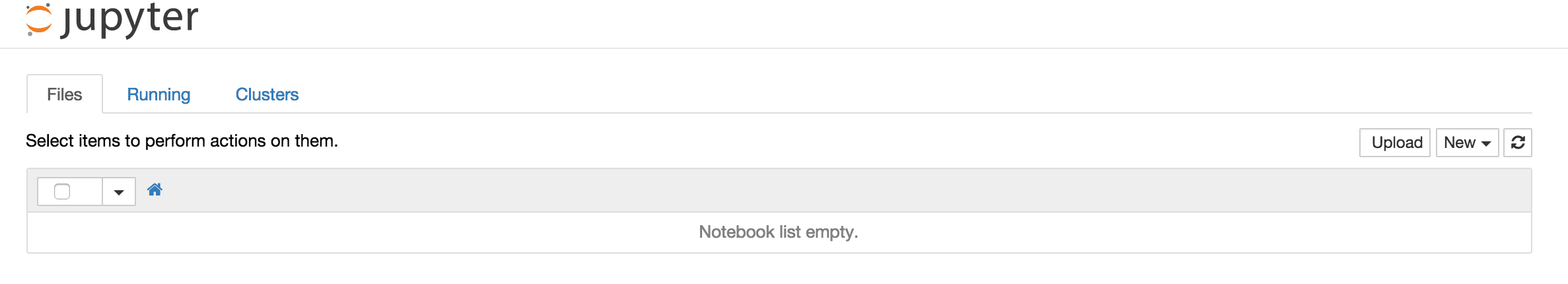
This is what you should see after navigating to the URL in your browser. If you see the screen above, you're successfully developing within the Docker container. To recap, instead of having to download Python, some runtime libraries, and the Jupyter package, all that was required was to install Docker, download the official Jupyter image, and run the container. Next, we'll expand on this and learn how to share notebooks from your host machine (local machine) with the running container.
Sharing Notebooks Between the Host and Container
To begin, we'll start by creating a directory on our host machine where we'll keep all of our notebooks. In your home directory, create a new directory called notebooks. Here's one command to do that: mkdir ~/notebooks
Now that we have a dedicated directory for our notebooks, we can share this directory between the host and container. Similar to opening the ports, we'll need to pass in another additional argument to the run command. The flag for this argument is -v <host_directory>:<container_directory> which tells the Docker engine to mount the given host directory to the container directory.
From the Jupyter Docker documentation, it specifies the working directory of the container as /home/jovyan. Thus, we'll mount our ~/notebooks directory to the container's working directory using the mount run flag.
docker run -p 8888:8888 -v ~/notebooks:/home/jovyan jupyter/minimal-notebookWith the directory mounted, go to the Jupyter server and create a new notebook. Rename the notebook from "Unititled" to "Example Notebook".

On your host machine, check the ~/notebooks directory. In there, you should see an iPython file: Example Notebook.ipynb!
Installing Additional Packages
In our minimal-notebook Docker image, there are pre-installed Python packages available for use. One of them we have been using explicitly, the Jupyter notebook, which is the notebook server that we're accessing on the browser. Other packages are implicitly installed, like the requests package, which you can import within a notebook. Notice that these pre-installed packages were bundled in the image, we did not install them ourselves.
As we have mentioned, not having to install packages is one of the major benefits of using Docker for development. But, what if an image is missing a data science package you wanted to use, say something like tensorflow for machine learning? One way to install a package in your container is to use the docker exec command.
The exec command has similar arguments with the run command, but it doesn't start a container with the arguments, it executes on an already running container. So, instead of creating a container from an image, like docker run, docker exec requires a running container ID or container name which are called container identifiers. To locate a running container's identifiers, you need to call the docker ps commandwhich lists all the running containers and some additional info. For example, here's what our docker ps outputs while the minimal-notebook container is running.
$ docker ps
CONTAINER ID IMAGE COMMAND CREATED STATUS PORTS NAMES
874108dfc9d9 jupyter/minimal-notebook "tini -- start-noteb…" Less than a second ago Up 4 seconds 0.0.0.0:8900->8888/tcp thirsty_almeidaNow that we have the container ID, we can install Python packages in the container. From the docker exec documentation, we pass in runnable command as an argument proceeding the identifier which is then executed in the container. Recall that the command to install Python packages is pip install <package name>.
To install tensorflow, we'll run the following in our shell. NB:, your container ID will be different from the provided example. docker exec 874108dfc9d9 pip install tensorflow
If successful, you should see the pip installation output logs. One thing you'll notice is that the installation of tensorflow within this Docker container is relatively quick (given you have fast internet). If you've ever installed tensorflow before, you'll know that it's quite the tedious setup, so you might be surprised at how painless this process is. The reason it is a quick installation process, is because the minimal-notebook image has been written with data science optimization in mind. The C libraries and other Linux system level packages have already been pre-installed based on the Jupyter community's thoughts on installation best practices. This is the greatest benefit of using open source community developer Docker images as they are commonly optimized for the type of developement work you will be doing.
Extended Docker Images
Up to this point, you've installed Docker, ran your first container, accessed a Dockerized Jupyter container, and installed tensorflow on a running container. Now, suppose you've finished your day working on the Jupyter container using the tensorflow library, and you want to shut the container down to reclaim processing speed and memory.
To stop the container, you can run either the docker stop or docker rm commands. The next day, you rerun the container again, and are ready to start hacking on your tensorflow work. However, when you go to run the notebook cells, you're blocked by an ImportError. How could this happen if you already had installed tensorflow the previous day?

The problem lies in the docker exec command. Recall that when you run exec, you are executing the given command to the running container. The container is just a running process of the image, where the image is the executable that contains all the pre-installed libraries. So, when you install tensorflow on the container, it is only installed for that specific instance.
Therefore, shutting down the container is deleting that instance from memory, and when you restart a new container from the image, only the libraries contained in the image will be available for use again. The only way to save tensorflow to the list of installed packages on an image is to either modify the original Dockerfile and build a new image, or to extend the minimal-container Dockerfile and build a new image from this new Dockerfile.
Unfortunately, each of these steps require an understanding of Dockerfiles, a Docker concept that we won't cover in detail in this tutorial. However, we are using the Jupyter community developed Docker images, so let's check if there is already a built Docker image with tensorflow. Looking at the Jupyter github repository again, we can see that there is a tensorflow notebook!
Not only tensorflow, but there are quite a few other options as well. The following tree diagram from their documentation describes the relationship extension between the Dockerfiles, and each available image in use. Because
tensorflow-notebook is extended from minimal-notebook, we can use the same docker run command from before and only change the name of the image. Here's how we can run a Dockerized Jupyter notebook server preinstalled with tensorflow: docker run -p 8888:8888 -v ~/notebooks:/home/jovyan jupyter/tensorflow-notebook On your browser, navigate to the running server using the same method described a few sections ago. There, run import tensorflow in a code cell and you should see no ImportError!
Next Steps
In this tutorial, we covered the differences between virtualization and containerization, how to install and run Dockerized applications, and the benefits of using open-source community developer Docker images. We used a containerized Jupyter notebook server as an example, and showed how painless working on a Jupyter server within a Docker container is. Finishing this tutorial, you should feel comfortable working with Jupyter community images, and be able to implement a Docker setup in your daily work.
While we covered a lot of Docker concepts, these were only the basics to help you get started. There is a lot more to learn about Docker and how powerful of a tool it can be. Mastering Docker will not only help you in your local development time, but can save time and money when working with teams of data scientists. If you enjoyed this example, here are some improvements you can add to help learn additional Docker concepts:
- Run the Jupyter server in detached mode as a background process.
- Name your running container to keep your processes clean.
- Create your own Dockerfile that extends the
minimal-notebookthat contains your necessary data science libraries. - Build an image from the extended Dockerfile.