Computer Vision in PyTorch (Part 2): Preparing Data, Training, and Evaluating Your CNN for Pneumonia Detection
In Part 1 of this tutorial series, we explored the fundamentals of Convolutional Neural Networks (CNNs) and built a complete CNN architecture using PyTorch for pneumonia detection in chest X-rays. We learned why CNNs excel at image tasks, examined each component in detail, and implemented a custom PneumoniaCNN class by taking an OOP approach and subclassing PyTorch's nn.Module class.
Now it's time to bring our model to life! In this tutorial, we'll complete our pneumonia detection system by:
- Preparing and preprocessing the chest X-ray dataset
- Training our CNN model with a complete training loop
- Evaluating model performance using metrics like precision, recall, and F1
- Interpreting evaluation results with a focus on visualizing predictions
- Addressing common CNN training issues like overfitting, underfitting, and class imbalance
By the end of this tutorial, you'll have transformed your CNN architecture into a working medical diagnostic tool and gained practical skills for implementing and evaluating deep learning models. Let's get started!

Prerequisites
Before proceeding, make sure you've:
- Read Computer Vision in PyTorch (Part 1): Building Your First CNN for Pneumonia Detection
- Installed PyTorch (follow PyTorch's official installation instructions)
- Reviewed fundamental deep learning concepts such as:
- Layers
- Activation functions
- Loss
- Optimization
Below are the modules, functions, and classes we’ll need for this tutorial. Be sure to install any libraries you’re missing after running this code:
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torch.utils.data import DataLoader, Dataset
import torchvision
import torchvision.transforms as transforms
from torchvision.datasets import ImageFolder
import matplotlib.pyplot as plt
import numpy as np
import tarfile
import os
import collections
import random
from sklearn.metrics import confusion_matrix, classification_report
from sklearn.model_selection import train_test_split
from PIL import Image
import seaborn as sns1. Preparing and Preprocessing the X-ray Image Dataset
We’ll start by preparing our chest X-ray dataset. The dataset contains X-ray images of lungs classified into two categories: NORMAL and PNEUMONIA.
Downloading and Extracting the Dataset
The data for this tutorial is available for download here as a compressed tar.gz file. After downloading, you'll need to extract it to access the images:
# Path to the downloaded tar.gz file
dataset_path = "xray_dataset.tar.gz" # If saved to your current directory
# Extract the dataset
with tarfile.open(dataset_path, "r:gz") as tar:
tar.extractall()
print("Dataset extracted successfully")After extraction, you should have this directory structure:
chest_xray/
├── test/
│ ├── NORMAL/
│ └── PNEUMONIA/
└── train/
├── NORMAL/
└── PNEUMONIA/Verifying Dataset Structure and File Counts
After extracting the dataset, it's good practice to verify the contents and get a count of the image files. This ensures we're working with the correct data and helps identify potential issues early on.
We'll create a small helper function to scan the train and test directories. This function will gather the file paths for all valid JPEG images and their corresponding class labels (0 for NORMAL, 1 for PNEUMONIA). Collecting these paths and labels now will also prepare us for the next step in preparing our data for training.
# Define base directories relative to your notebook/script location
data_dir = "chest_xray"
train_dir = os.path.join(data_dir, "train")
test_dir = os.path.join(data_dir, "test")
# Define the classes based on the subfolder names
class_names = ['NORMAL', 'PNEUMONIA']
class_to_idx = {cls_name: i for i, cls_name in enumerate(class_names)}
# Helper function to scan directories, filter JPEG images, and collect paths/labels
def get_image_paths_and_labels(data_dir):
image_paths = []
labels = []
print(f"Scanning directory: {data_dir}")
for label_name in class_names:
class_dir = os.path.join(data_dir, label_name)
count = 0
# List files in the class directory
for filename in os.listdir(class_dir):
# Keep only files ending with .jpeg (case-insensitive)
if filename.lower().endswith('.jpeg'):
image_paths.append(os.path.join(class_dir, filename))
labels.append(class_to_idx[label_name])
count += 1
print(f" Found {count} '.jpeg' images for class '{label_name}'")
return image_paths, labels
# Get paths and labels for the training set
all_train_paths, all_train_labels = get_image_paths_and_labels(train_dir)
train_counts = collections.Counter(all_train_labels)
total_train_images = len(all_train_paths)
print(f"\nTraining Set Counts:")
print(f" NORMAL (Class 0): {train_counts[class_to_idx['NORMAL']]}")
print(f" PNEUMONIA (Class 1): {train_counts[class_to_idx['PNEUMONIA']]}")
print(f" Total Training Samples: {total_train_images}")
# Get paths and labels for the test set
all_test_paths, all_test_labels = get_image_paths_and_labels(test_dir)
test_counts = collections.Counter(all_test_labels)
total_test_images = len(all_test_paths)
print(f"\nTest Set Counts:")
print(f" NORMAL (Class 0): {test_counts[class_to_idx['NORMAL']]}")
print(f" PNEUMONIA (Class 1): {test_counts[class_to_idx['PNEUMONIA']]}")
print(f" Total Test Samples: {total_test_images}")Running this code will scan the directories and produce the following counts:
Scanning directory: chest_xray/train
Found 1349 '.jpeg' images for class 'NORMAL'
Found 3883 '.jpeg' images for class 'PNEUMONIA'
Training Set Counts:
NORMAL (Class 0): 1349
PNEUMONIA (Class 1): 3883
Total Training Samples: 5232
Scanning directory: chest_xray/test
Found 234 '.jpeg' images for class 'NORMAL'
Found 390 '.jpeg' images for class 'PNEUMONIA'
Test Set Counts:
NORMAL (Class 0): 234
PNEUMONIA (Class 1): 390
Total Test Samples: 624Excellent! Our helper function scanned the directories and gave us clean lists of all the usable JPEG images and their corresponding labels for both training and testing (stored in variables like all_train_paths, all_train_labels, etc.).
Now, looking at the training counts (1349 NORMAL to 3883 PNEUMONIA), something immediately stands out: there are almost three times as many pneumonia examples in our training data! This situation, where one class significantly outnumbers another, is called class imbalance.
While techniques exist to directly address class imbalance during training (we’ll talk about those later), our plan for now is to first train the model using the data as-is. That said, this imbalance means we'll need to be especially careful when we get to evaluating the model's performance. We can't just rely on overall accuracy; we'll need to use specific metrics that tell us how well the model identifies both classes fairly. But we’ll get to that too.
Having prepared the lists of training image paths and labels, we're now ready for the next important step in preparing our data: splitting off a portion of the training images to create a validation set.
Why You Need a Validation Set
Now that we have lists of our training images and associated labels, you might be thinking, “Why did we need those specific lists?” Well, before we train our model, we need to set aside a portion of that training data to create a validation set.
It might seem strange to not use all available data for training, but this validation split is vital for trustworthy model development. Here's why:
- Tuning & Monitoring: While training, we need to monitor how well the model is learning and potentially tune things like the learning rate or decide when to stop training. We need a dataset for this that the model isn't directly training on, but which isn't our final, untouched test set. That's the validation set's job.
- Avoiding Data Leakage: If we used the test set to make these tuning decisions, we'd essentially be "leaking" information about the test set into our model development process. The model might end up looking good on that specific test set simply because we optimized for it, but fail to generalize to new, truly unseen data.
- Unbiased Final Test: The test set should only be used once, at the very end, after all training and tuning are complete, to get an unbiased estimate of the final model's performance.
So, we reserve the test_paths/test_labels for the final evaluation and split our all_train_paths/all_train_labels into two new subsets: one for actual training, and one for validation during development.
We'll use the train_test_split function from scikit-learn for this. Because we identified a class imbalance earlier, we'll use the stratify option to ensure both the new training subset and the validation set maintain the original proportion of NORMAL and PNEUMONIA images. Using random_state ensures the split is the same every time the code runs.
# Define proportion for validation set
val_split_ratio = 0.2
SEED = 42
# Perform stratified split
train_paths, val_paths, train_labels, val_labels = train_test_split(
all_train_paths,
all_train_labels,
test_size=val_split_ratio,
stratify=all_train_labels,
random_state=SEED
)
# Print the number of samples in each resulting set
print(f"Original training image count: {len(all_train_paths)}")
print(f"--> Split into {len(train_paths)} training samples")
print(f"--> Split into {len(val_paths)} validation samples")Running this will perform the split and show the resulting counts:
Original training image count: 5232
--> Split into 4185 training samples
--> Split into 1047 validation samplesWe now have distinct lists of file paths and corresponding labels for our training data (train_paths, train_labels) and our validation data (val_paths, val_labels). These lists tell us which images belong in each set.
But simply having file paths isn't enough to feed data into a PyTorch model. Each image needs to be loaded and undergo several processing steps first. These include standard operations like resizing all images to a consistent 256×256 dimension and converting them into the correct format (single-channel grayscale tensors). Additionally, to help our model learn more robust features from a smaller dataset and generalize better from our specific training images, we'll apply a technique called data augmentation, but only on the training set.
Understanding Data Augmentation
We have our training images identified, but deep learning models often benefit from seeing a large variety and quantity of data. What if our training set, particularly after splitting, isn't large or diverse enough to teach the model to generalize well to all possible variations it might encounter in new X-rays? This is where data augmentation comes in.
What is Data Augmentation?
Data augmentation is a technique used to artificially increase the diversity of your training dataset without actually collecting new images. It involves applying random, yet realistic, transformations to the images during the training process. Each time the model sees an image from the training set, it might see a slightly altered version (e.g., flipped horizontally or slightly rotated).
Why Use Data Augmentation?
- Improved Generalization & Robustness: By exposing the model to these variations (like different orientations or flips), it learns to focus on the underlying patterns relevant to the task (e.g., signs of pneumonia) rather than potentially irrelevant characteristics like the exact positioning of the patient. This helps the model generalize better to new, unseen images that might have similar slight variations.
- Reduced Overfitting: It effectively increases the perceived size of the training set, making it harder for the model to simply memorize the training examples. This is particularly valuable when working with specialized datasets (like medical images) that might be smaller than general-purpose image datasets.
Our Chosen Augmentations:
For this tutorial, we'll apply two simple and common augmentation techniques using torchvision.transforms:
transforms.RandomHorizontalFlip(0.5): This randomly flips the image horizontally (left-to-right) with a default probability of 50%.transforms.RandomRotation(10): This randomly rotates the image by a small angle, in this case, up to 10 degrees in either direction.
These simple variations help the model learn features that aren't dependent on perfect orientation or specific left-right positioning. Many other augmentation techniques exist, like adjusting brightness/contrast, zooming, or shearing, but we'll stick to these two for now.
Important: Training Only!
Crucially, data augmentation is applied only to the training set. We do not apply random augmentations to the validation or test sets. Why? Because we need a consistent and unbiased measure of the model's performance on unmodified data during validation (for tuning) and testing (for final evaluation). Augmenting validation/test data would introduce randomness that makes performance measurement unreliable.
Now that we understand the concept and benefits of data augmentation, let's define the complete image transformation pipelines for our training, validation, and test sets.
Defining Image Transforms
Now that we have lists specifying which images belong to our training and validation sets, we need to define how to process each image file into a standardized tensor format suitable for our PyTorch model. This involves creating processing pipelines using torchvision.transforms. We'll need slightly different pipelines for training data (which includes random augmentation) and for validation/test data (which does not).
These pipelines need to perform several key operations consistently for every image:
- Resize to Fixed Dimensions: To meet our model's required 256×256 input size, the first step is
transforms.Resize((256, 256)). Be aware that because the original X-rays vary in size, this forces a square aspect ratio and will distort non-square images by stretching or squashing them. While this could potentially obscure subtle diagnostic cues related to shape or proportion, CNNs can often adapt and learn effectively from such consistently distorted data. We'll use this standard resizing approach for our fixed-input model, but keep in mind that if evaluation reveals performance issues potentially linked to shape distortion, exploring aspect-preserving alternatives (like padding before resizing) would be a logical next step to investigate. - Ensure Grayscale: The model architecture also expects single-channel grayscale images (
in_channels=1). To guarantee this format for all images processed, we includetransforms.Grayscale(num_output_channels=1). - Data Augmentation (Training Only): For the training pipeline (
train_transforms), we'll insert thetransforms.RandomHorizontalFlip()andtransforms.RandomRotation(10)steps discussed in the previous section to help the model generalize better. These are not included in the validation/test pipeline. - Convert to Tensor & Scale: The final step is
transforms.ToTensor(). This performs two critical functions: it converts the processed PIL Image object into a PyTorch tensor, and it scales the pixel values from the original integer range [0, 255] down to a floating-point range of [0.0, 1.0]. This [0, 1] scaling acts as our input normalization for this tutorial. We are opting for this simpler approach instead of standardizing with a separatetransforms.Normalize(mean, std)step, relying partly on theBatchNorm2dlayers within our model to help adapt to the input distribution during training.
Creating the Pipelines
With these steps decided, we define two distinct data process pipelines using transforms.Compose:
# Transformations for the training set (including augmentation)
train_transforms = transforms.Compose([
transforms.Resize((256, 256)),
transforms.Grayscale(num_output_channels=1),
transforms.RandomHorizontalFlip(),
transforms.RandomRotation(10),
transforms.ToTensor() # Converts to tensor AND scales to [0, 1]
])
# Transformations for the validation and test sets (NO augmentation)
val_test_transforms = transforms.Compose([
transforms.Resize((256, 256)),
transforms.Grayscale(num_output_channels=1),
transforms.ToTensor() # Converts to tensor AND scales to [0, 1]
])
print("Transformation pipelines defined.")Now that we've defined how to process the images with train_transforms and val_test_transforms, we need an efficient way to connect these pipelines to our lists of image paths (train_paths, val_paths, etc.). Specifically, we need a structure that can take an index, find the corresponding image path and label, load the image file, apply the correct transformations, and provide the resulting tensor and label to PyTorch for training or evaluation.
This requires creating a custom PyTorch Dataset. Let's build that next.
Creating a Custom PyTorch Dataset
We have our lists of image paths, and we have our processing pipelines, so now let’s bring them together so PyTorch can load and transform images during training and evaluation.
While PyTorch offers built-in datasets like ImageFolder, they assume a specific directory structure and aren't ideal for using pre-split lists of file paths with different transforms assigned to each split. Thankfully, PyTorch makes it straightforward to create our own custom dataset handling logic by inheriting from the base torch.utils.data.Dataset class.
A custom Dataset needs to implement three essential methods:
__init__(self, ...): Initializes the dataset, typically by storing file paths, labels, and any necessary transformations.__len__(self): Returns the total number of samples in the dataset.__getitem__(self, idx): Loads and returns a single sample (usually an image tensor and its label) from the dataset, given an indexidx. This method is where the image loading and transformations are actually applied, often "just-in-time" when the sample is requested.
Let's define our XRayDataset class:
class XRayDataset(Dataset):
"""Custom Dataset for loading X-ray images from file paths."""
def __init__(self, image_paths, labels, transform=None):
"""
Args:
image_paths (list): List of paths to images.
labels (list): List of corresponding labels (0 or 1).
transform (callable, optional): Optional transform to be applied
on a sample.
"""
self.image_paths = image_paths
self.labels = labels
self.transform = transform
def __len__(self):
"""Returns the total number of samples in the dataset."""
return len(self.image_paths)
def __getitem__(self, idx):
"""
Fetches the sample at the given index, loads the image,
applies transformations, and handles potential errors.
Args:
idx (int): The index of the sample to fetch.
Returns:
tuple: (image_tensor, label) if successful.
None: If an error occurs (e.g., file not found, processing error),
signalling to skip this sample.
"""
# Get the path and label for the requested index
img_path = self.image_paths[idx]
label = self.labels[idx]
try:
# Load the image using PIL within a context manager
with Image.open(img_path) as img:
# Apply transforms ONLY if they exist
if self.transform:
# Apply the entire transform pipeline
image_tensor = self.transform(img)
# Return the processed tensor and label
return image_tensor, label
else:
# This branch indicates a setup error, as the transform
# pipeline should at least contain ToTensor().
raise ValueError(f"Dataset initialized without transforms for {img_path}. "
"Transforms (including ToTensor) are required.")
except FileNotFoundError:
# Handle cases where the image file doesn't exist
print(f"Warning: Image file not found at {img_path}. Skipping sample {idx}.")
return None # Returning None signals to skip
except ValueError as e:
# Catch the specific error we raised for missing transforms
print(f"Error for sample {idx} at {img_path}: {e}")
raise e # Re-raise critical setup errors
except Exception as e:
# Catch any other PIL loading or transform errors
print(f"Warning: Error processing image {img_path} (sample {idx}): {e}. Skipping sample.")
return None # Returning None signals to skipExplanation:
__init__: The constructor (__init__) is straightforward. It simply stores the essential information passed when we create an instance ofXRayDataset: the list of image paths, the corresponding list of labels, and the specifictorchvision.transformspipeline that should be applied to images from this dataset.__len__: This method allows PyTorch code to easily get the total size of the dataset by simply returning the number of image paths provided during initialization.__getitem__: This is the core method where the actual data loading and processing happens for a single sample. When requested by its index (idx), it performs the following steps:- Retrieves the image file path and label using the index.
- Opens the image file using the PIL library.
- Applies the entire transformation pipeline (like
train_transformsorval_test_transforms) stored inself.transform. - Returns the processed image tensor and its integer label if successful.
Crucially, this loading and transforming happens "on demand" or "lazily." The implementation also includes basic error handling: if an image file is missing or fails during processing, it prints a warning and returnsNone, signaling that this sample should be skipped.
This XRayDataset class gives us a blueprint for handling our image data. With this class defined, we can now create the specific Dataset instances we need: one for our training data using train_paths and train_transforms, one for validation using val_paths and val_test_transforms, and one for our test set. Let's instantiate these datasets next.
Creating Final Datasets and DataLoader Objects
With our XRayDataset class ready, we can now instantiate it for each of our data splits. We'll pair the appropriate lists of image paths and labels with the corresponding transformation pipelines we defined earlier.
# Instantiate the custom Dataset for each split
train_dataset = XRayDataset(
image_paths=train_paths,
labels=train_labels,
transform=train_transforms # Apply training transforms (incl. augmentation)
)
val_dataset = XRayDataset(
image_paths=val_paths,
labels=val_labels,
transform=val_test_transforms # Apply validation transforms (no augmentation)
)
test_dataset = XRayDataset(
image_paths=all_test_paths, # Using all_test_paths from verification step
labels=all_test_labels, # Using all_test_labels from verification step
transform=val_test_transforms # Apply validation/test transforms
)
# Print dataset sizes to confirm
print("\nFinal Dataset objects created:")
print(f" Training dataset size: {len(train_dataset)}")
print(f" Validation dataset size: {len(val_dataset)}")
print(f" Test dataset size: {len(test_dataset)}")This gives us three Dataset objects, each knowing how to access and transform its specific set of images.
Final Dataset objects created:
Training dataset size: 4185
Validation dataset size: 1047
Test dataset size: 624Introducing DataLoader
While Dataset objects allow us to access individual processed samples via dataset[index], we typically train neural networks on mini-batches of data, not one sample at a time. Processing batches is more computationally efficient and helps stabilize the learning process.
PyTorch's torch.utils.data.DataLoader class is designed precisely for this. It takes a Dataset object and provides an iterable that yields batches of data. Key features include:
- Batching: Automatically groups individual samples from the
Datasetinto batches of a specified size (batch_size). - Shuffling: Can automatically shuffle the training data at the beginning of each epoch (
shuffle=True) to ensure the model doesn't learn based on the order of examples. Shuffling is typically disabled for validation and testing for consistent evaluation. - Parallel Loading: Can use multiple background worker processes (
num_workers) to load data concurrently, preventing data loading from becoming a bottleneck during training, especially when using a GPU. Thenum_workersargument specifies how many subprocesses to use for data loading. While values > 0 can speed things up by loading data in parallel, they can sometimes cause issues in certain environments (like Colab notebooks). If you encounter errors during training related to workers, try settingnum_workers=0, which loads data in the main process. - Memory Pinning: Can use
pin_memory=Trueto speed up data transfer from CPU to GPU memory when training on CUDA-enabled devices.
Creating the DataLoader Instances
Let's create DataLoader instances for each of our datasets:
# Define batch size (can be tuned depending on GPU memory)
batch_size = 32
# Create DataLoader for the training set
train_loader = DataLoader(
dataset=train_dataset,
batch_size=batch_size,
shuffle=True, # Shuffle data each epoch for training
num_workers=2, # Number of subprocesses to use for data loading (adjust based on system)
pin_memory=True # Speeds up CPU-GPU transfer if using CUDA
)
# Create DataLoader for the validation set
val_loader = DataLoader(
dataset=val_dataset,
batch_size=batch_size,
shuffle=False, # No need to shuffle validation data
num_workers=2,
pin_memory=True
)
# Create DataLoader for the test set
test_loader = DataLoader(
dataset=test_dataset,
batch_size=batch_size,
shuffle=False, # No need to shuffle test data
num_workers=2,
pin_memory=True
)
print(f"\nDataLoaders created with batch size {batch_size}.")With train_loader, val_loader, and test_loader created, our data preparation pipeline is complete! These loaders are now ready to efficiently supply batches of preprocessed image tensors and labels to our model during the training, validation, and testing phases.
A good next step is often to visualize a few images from the train_loader to visually inspect the results of the transformations and augmentations before proceeding to model training.
Visualizing Sample Images
Before we start training, it's crucial to visually inspect the output of our DataLoader objects. This acts as a sanity check to ensure our data loading, preprocessing, and augmentation steps are working correctly – essentially, we get to "see what the model will see."
Let's create a helper function to display a batch of images:
def show_batch(dataloader, class_names, title="Sample Batch", n_samples=8):
"""Displays a batch of transformed images from a DataLoader."""
try:
images, labels = next(iter(dataloader)) # Get one batch
except StopIteration:
print("DataLoader is empty or exhausted.")
return
# Limit number of samples to display if batch is smaller than n_samples
actual_samples = min(n_samples, images.size(0))
if actual_samples <= 0:
print("No samples found in the batch to display.")
return
images = images[:actual_samples]
labels = labels[:actual_samples]
# Tensors are likely on GPU if device='cuda', move to CPU for numpy/plotting
images = images.cpu()
labels = labels.cpu()
# Determine subplot layout
if actual_samples <= 4:
ncols = actual_samples; nrows = 1; figsize = (3 * ncols, 4)
else:
ncols = 4; nrows = 2; figsize = (12, 6)
fig, axes = plt.subplots(nrows=nrows, ncols=ncols, figsize=figsize)
if nrows == 1 and ncols == 1: axes = np.array([axes]) # Handle single plot case
axes = axes.flatten() # Flatten axes array for easy iteration
fig.suptitle(title, fontsize=16)
for i in range(actual_samples):
ax = axes[i]
img_tensor = images[i] # Shape is [C=1, H, W], scaled [0.0, 1.0]
# Reminder: ToTensor scaled pixels to [0, 1]
# Matplotlib can directly display tensors in this range with cmap='gray'
# Permute dimensions from [C, H, W] to [H, W, C] for matplotlib
img_display = img_tensor.permute(1, 2, 0).numpy()
# Display the image, removing the channel dimension using squeeze() for grayscale
# Specify vmin/vmax ensures correct display range for float data
ax.imshow(img_display.squeeze(), cmap='gray', vmin=0.0, vmax=1.0)
ax.set_title(f"Class: {class_names[labels[i]]}") # Use passed class_names
ax.axis('off')
# Hide any unused subplots if the grid is larger than needed
for j in range(actual_samples, len(axes)):
axes[j].axis('off')
plt.tight_layout()
plt.subplots_adjust(top=0.88 if title else 0.95, hspace=0.3) # Adjust for suptitle
plt.show()
# Visualize training samples (should show augmentations)
print("\nVisualizing a batch from train_loader...")
show_batch(train_loader, class_names, title="Sample Processed Training Images")
# Visualize validation samples (should NOT show augmentations)
print("\nVisualizing a batch from val_loader...")
show_batch(val_loader, class_names, title="Sample Processed Validation Images")Visualizing a batch from train_loader...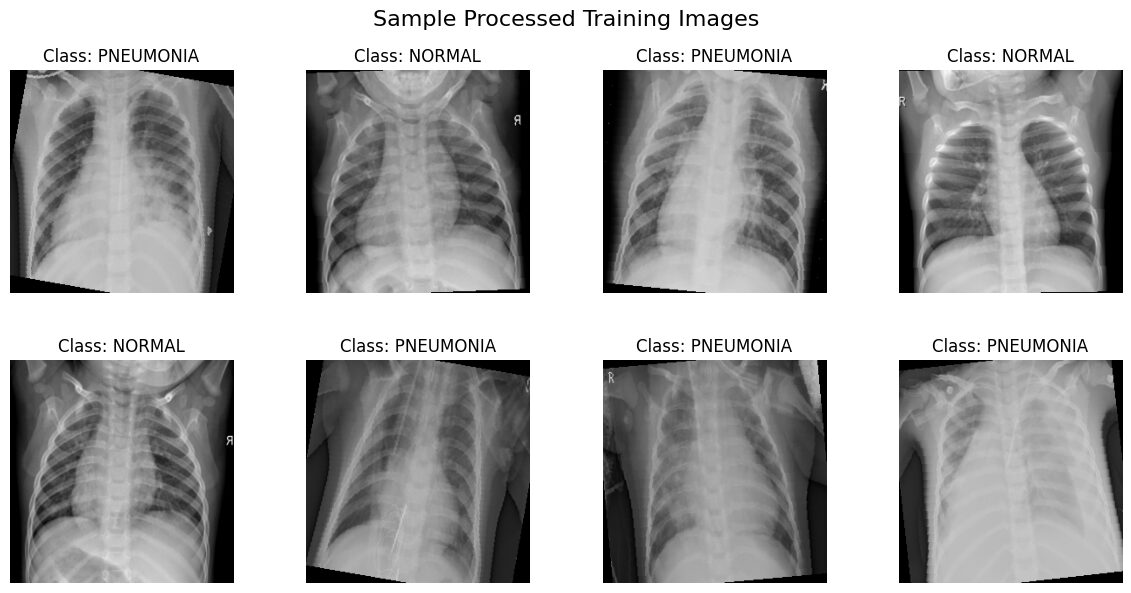
Visualizing a batch from val_loader...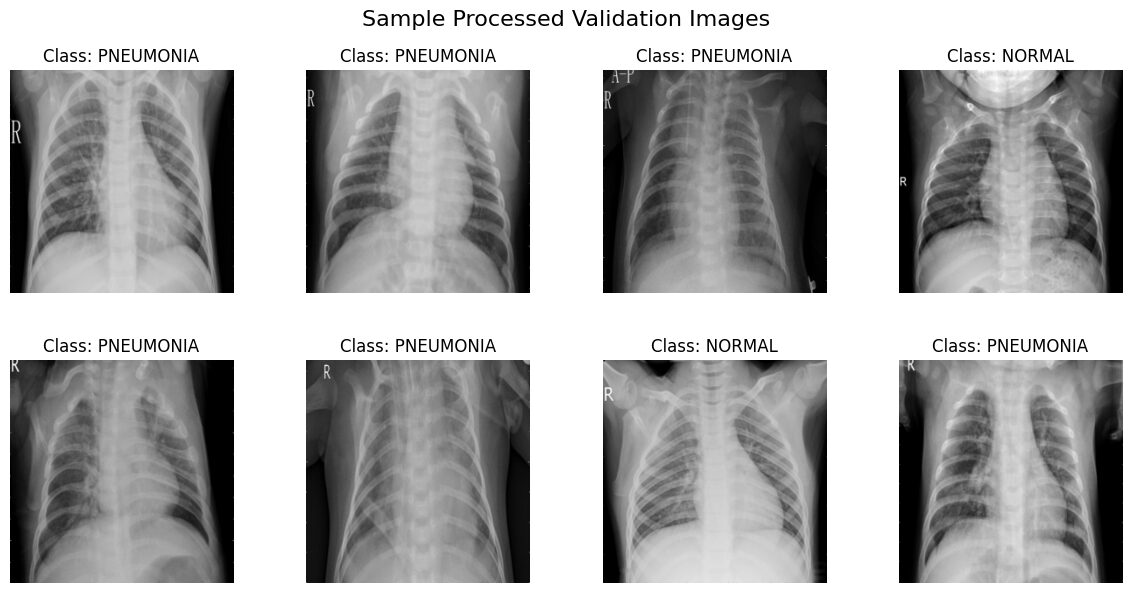
Interpreting the Visualizations
The images displayed above are samples drawn directly from our train_loader and val_loader. They reflect the full preprocessing pipeline:
- Resized to 256×256 pixels.
- Converted to single-channel grayscale.
- If from
train_loader: Randomly flipped horizontally and/or slightly rotated due to data augmentation. - Converted to PyTorch tensors with pixel values scaled to the [0.0, 1.0] range via
transforms.ToTensor.
What to Look For:
- Format: You should see grayscale chest X-rays, all uniformly sized.
- Labels: Each image should have the correct class title ('NORMAL' or 'PNEUMONIA').
- Augmentation: Images from
train_loadermight show random variations (flips, rotations) each time you run this visualization. Images fromval_loadershould appear consistent without these random effects. - Intensity Range: The images are displayed directly from the [0, 1] scaled tensors. Ensure they look reasonable (not all black or all white, details visible).
- Orientation Marker & Augmentation: You'll likely notice a letter marker, commonly an 'R', often placed in an upper corner of the X-rays. This marker indicates the patient's right side. Since standard chest X-rays are taken with the patient facing the detector, their right side appears on the left side of the image. Now, look closely at the samples from the
train_loader: ifRandomHorizontalFlipwas applied to an image, you’ll see this 'R' marker appearing reversed and on the right side of the image! This is a perfect visual confirmation that your training data augmentation is active. Images from theval_loadershould consistently show the marker in its standard position (patient's right on the image's left).
This visualization step confirms that our data loaders are correctly yielding processed image tensors in the format and range our model expects, with augmentations applied appropriately. With this confirmation, our data is ready for the main event: training the CNN model.
2. Training Our CNN Model
With our data prepared and loaded efficiently using DataLoader objects, we're ready to move on to the main event: training the model to distinguish between NORMAL and PNEUMONIA chest X-rays. For this, we'll use the PneumoniaCNN architecture we carefully designed together previously.
Instantiating the Model and Setting the Device
The first step is to use the PneumoniaCNN class definition we built in Part 1 of this tutorial series. You'll need to make sure that Python class definition is available in your current environment, typically by copying the class PneumoniaCNN(nn.Module): ... block into a code cell and running it here if you haven't already.
Once the PneumoniaCNN class is defined, we can create an instance of it. Don’t forget that we must then immediately move this model instance to the appropriate computing device (cpu or cuda) that we set up earlier. Performing operations between the model and data requires them both to reside on the same device.
# Instantiate the model
model = PneumoniaCNN()
# Check if CUDA (GPU support) is available, otherwise use CPU
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# Move the model to the chosen device (GPU or CPU)
model.to(device)
print(f"Model '{type(model).__name__}' instantiated and moved to '{device}'.")Now our model object is created and resides on the correct device. Before we can start the training loop itself, we need two more key components:
- A Loss Function: To measure how inaccurate the model's predictions are compared to the true labels.
- An Optimizer: To define the algorithm used to update the model's weights based on the calculated loss.
Let's define these next.
Defining the Loss Function and Optimizer
With our model instantiated and placed on the correct device, we need two final components before building the training loop:
- Loss Function: This measures how far the model's predictions (logits) are from the actual target labels. The computed loss value is what the model tries to minimize during training.
- Optimizer: This implements an algorithm (like Stochastic Gradient Descent or variations thereof) that updates the model's weights based on the gradients computed during the backward pass, aiming to reduce the loss.
Let's define these for our task:
# Define loss function
criterion = nn.CrossEntropyLoss()
# Define optimizer
optimizer = optim.Adam(model.parameters(), lr=0.0001)
print("Loss function and optimizer defined.")Explanation:
- Loss Function (
criterion): We instantiatenn.CrossEntropyLoss. This is the standard choice for multi-class classification problems like ours (Normal vs. Pneumonia). It's particularly convenient because it expects the raw, unnormalized scores (logits) directly from the model's final layer and internally applies the necessary calculations (like LogSoftmax and Negative Log-Likelihood loss) to determine the error. - Optimizer (
optimizer): We selectoptim.Adam, a very popular and often effective optimization algorithm. It's known for its adaptive learning rate capabilities, meaning it can adjust the learning rate for each parameter during training, which frequently leads to faster convergence compared to simpler optimizers like basic SGD.model.parameters(): We pass this to the optimizer to tell it exactly which tensors within ourmodelare the learnable weights and biases that it should be updating.lr=0.0001: This argument sets the initial learning rate. It's a crucial hyperparameter controlling how large the updates to the weights are on each step. A value between0.001and0.0001is often a good starting point for the Adam optimizer, but it might need tuning later.
Alright, all the preparatory pieces are in place! We have our instantiated model ready on the correct device, our DataLoaders (train_loader, val_loader, test_loader) prepared to serve batches of processed data, our criterion defined to measure loss, and our optimizer configured to update the model's parameters. We're finally ready to orchestrate the actual learning process by implementing the training loop.
Implementing the Training Loop
Now let's implement a complete training loop:
def train_model(model, train_loader, val_loader, criterion, optimizer, device, num_epochs=20):
"""Trains and validates the model."""
# Initialize lists to track metrics
train_losses = []
val_losses = []
train_accuracies = []
val_accuracies = []
print("Starting Training...")
# Training loop
for epoch in range(num_epochs):
# Training Phase
model.train() # Set model to training mode (enables dropout, batch norm updates)
running_loss = 0.0
correct_train = 0
total_train = 0
# Iterate over training data
for i, (images, labels) in enumerate(train_loader):
# Move data to the specified device
images, labels = images.to(device), labels.to(device)
# Zero the parameter gradients
optimizer.zero_grad()
# Forward pass
outputs = model(images)
loss = criterion(outputs, labels)
# Backward pass and optimize
loss.backward()
optimizer.step()
# Track training loss and accuracy
running_loss += loss.item() * images.size(0) # loss.item() is the avg loss per batch
_, predicted = torch.max(outputs.data, 1)
total_train += labels.size(0)
correct_train += (predicted == labels).sum().item()
# Calculate training statistics for the epoch
epoch_train_loss = running_loss / len(train_loader.dataset)
epoch_train_acc = correct_train / total_train
train_losses.append(epoch_train_loss)
train_accuracies.append(epoch_train_acc)
# Validation Phase
model.eval() # Set model to evaluation mode (disables dropout, uses running stats for batch norm)
val_loss = 0.0
correct_val = 0
total_val = 0
# Disable gradient calculations for validation
with torch.no_grad():
for images, labels in val_loader:
images, labels = images.to(device), labels.to(device)
outputs = model(images)
loss = criterion(outputs, labels)
val_loss += loss.item() * images.size(0)
_, predicted = torch.max(outputs.data, 1)
total_val += labels.size(0)
correct_val += (predicted == labels).sum().item()
# Calculate validation statistics for the epoch
epoch_val_loss = val_loss / len(val_loader.dataset)
epoch_val_acc = correct_val / total_val
val_losses.append(epoch_val_loss)
val_accuracies.append(epoch_val_acc)
# Print statistics for the epoch
print(f"Epoch {epoch+1}/{num_epochs}")
print(f" Train Loss: {epoch_train_loss:.4f}, Train Acc: {epoch_train_acc:.4f}")
print(f" Val Loss: {epoch_val_loss:.4f}, Val Acc: {epoch_val_acc:.4f}")
print("-" * 30)
print("Finished Training.")
# Return performance history
return {
'train_losses': train_losses,
'train_accuracies': train_accuracies,
'val_losses': val_losses,
'val_accuracies': val_accuracies
}This training function:
- Tracks performance metrics (training and validation) over time.
- Switches between training (
model.train()) and evaluation (model.eval()) modes correctly. - Handles device placement for tensors (
.to(device)). - Implements the full train-validate cycle for each epoch.
- Returns a dictionary of training and validation history for later analysis.
Now, let's start training our model:
# Train the model
num_epochs = 20
history = train_model(
model=model,
train_loader=train_loader,
val_loader=val_loader,
criterion=criterion,
optimizer=optimizer,
device=device,
num_epochs=num_epochs
)During training, you'll see output showing the model's progress:
Starting Training...
Epoch 1/20
Train Loss: 0.5181, Train Acc: 0.8282
Val Loss: 0.1428, Val Acc: 0.9484
------------------------------
Epoch 2/20
Train Loss: 0.2066, Train Acc: 0.9221
Val Loss: 0.0897, Val Acc: 0.9685
------------------------------
Epoch 3/20
Train Loss: 0.1632, Train Acc: 0.9379
Val Loss: 0.0708, Val Acc: 0.9780
------------------------------
... (output for subsequent epochs) ...
------------------------------
Epoch 20/20
Train Loss: 0.0832, Train Acc: 0.9699
Val Loss: 0.0468, Val Acc: 0.9819
Finished Training.Training is complete! The output above gives us a snapshot of the loss and accuracy progress for each epoch on both the training and validation sets. We can see the model is learning, but to get a full picture of the trends over all 20 epochs, like how quickly the model converged, whether overfitting occurred, and how the validation performance truly compared to training, we should visualize these metrics instead. So let's plot the history next.
Visualizing the Training Process
Visualizing the training and validation metrics is the best way to understand how our model is learning. Plotting their loss/accuracy curves over epochs provides valuable insights into the learning dynamics.
def plot_training_history(history):
"""Plots the training and validation loss and accuracy."""
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(15, 5))
# Plot losses
ax1.plot(history['train_losses'], label='Train Loss')
ax1.plot(history['val_losses'], label='Validation Loss')
ax1.set_xlabel('Epoch')
ax1.set_ylabel('Loss')
ax1.set_title('Training and Validation Loss')
ax1.legend()
ax1.grid(True)
# Plot accuracies
ax2.plot(history['train_accuracies'], label='Train Accuracy')
ax2.plot(history['val_accuracies'], label='Validation Accuracy')
ax2.set_xlabel('Epoch')
ax2.set_ylabel('Accuracy')
ax2.set_title('Training and Validation Accuracy')
ax2.legend()
ax2.grid(True)
plt.tight_layout()
plt.show()
# Plot the training and validation history
plot_training_history(history)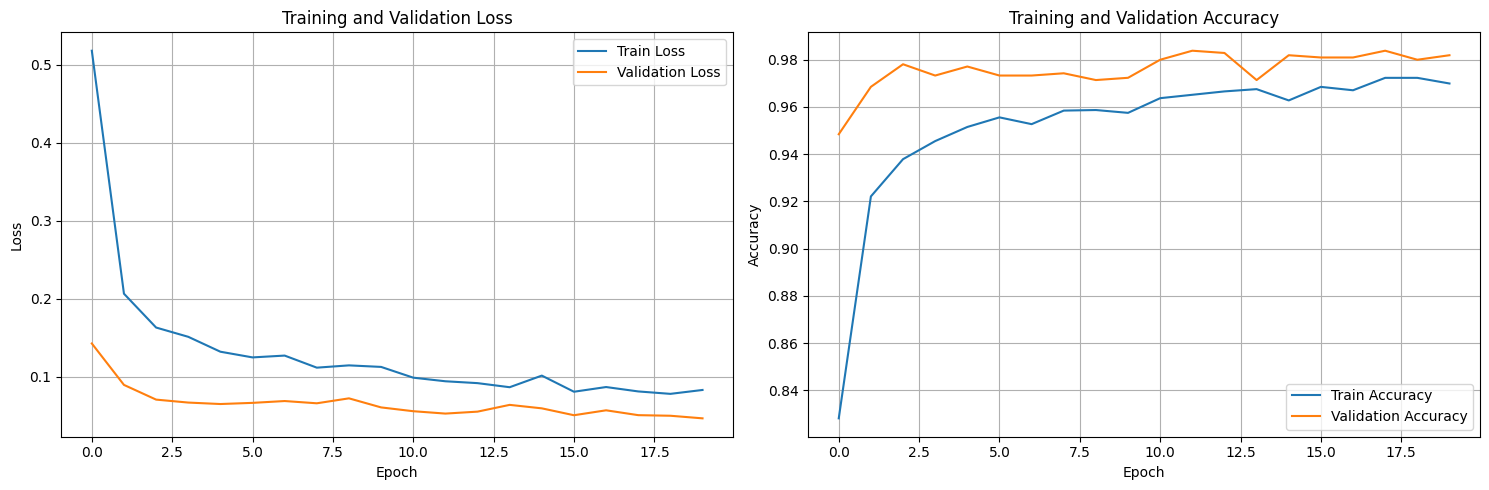
These plots provide a clear visual summary of the entire training process over the 20 epochs.
- Overall Learning: We can clearly see the learning trend: both the blue (training) and orange (validation) loss curves decrease significantly from the start and then begin to level off, particularly towards the end. Correspondingly, both accuracy curves rise quickly and plateau at a high level. This confirms the model successfully learned from the data.
- Validation vs. Training Performance: Notice how the orange validation loss curve consistently stays below the blue training loss curve, and the orange validation accuracy curve stays above the blue training accuracy curve. This pattern, where validation metrics appear better than training metrics, is often expected when using regularization techniques like Data Augmentation and Dropout. These techniques are applied only during the training phase (
model.train()), making that phase slightly harder, but are turned off during validation (model.eval()), allowing the model's full capacity to be assessed on the consistent validation data. - Overfitting Check: We visually inspect the gap between the training and validation curves. Signs of significant overfitting would include the validation loss (orange) clearly starting to rise while the training loss (blue) continues to fall, or the validation accuracy stalling/dropping while training accuracy keeps climbing. Based on these plots, while there are minor fluctuations, the validation loss remains low and generally trends downwards or flat near the end. The gap between the curves doesn't appear to be dramatically widening, suggesting significant overfitting hasn't set in within these 20 epochs.
- Optimal Epoch & Training Duration: Looking closely at the orange validation loss curve, it appears to reach its minimum value very late in training, around epoch 19 or 20. Similarly, validation accuracy plateaus at its peak in the last few epochs. This suggests that training for the full 20 epochs was beneficial for this specific run and learning rate, and stopping much earlier might have resulted in slightly suboptimal validation performance.
TL;DR: The plots show stable training with good convergence over 20 epochs. They visually confirm the expected impact of our training-only regularization (Val > Train metrics) and indicate that the model reached its best validation performance near the end of this training run without showing strong signs of overfitting yet.
3. Evaluating Our Pneumonia Detection CNN
After training, we need to rigorously evaluate our model to understand its strengths and weaknesses, especially for a medical application like pneumonia detection.
Calculating Key Metrics
For medical diagnosis tasks, accuracy alone is insufficient. We need to consider:
- Precision: Of all cases predicted as pneumonia, how many actually have pneumonia?
- Recall: Of all actual pneumonia cases, how many did we correctly identify?
- F1-score: The harmonic mean of precision and recall
- Confusion Matrix: A table showing true positives, false positives, true negatives, and false negatives
Let's implement a detailed evaluation:
def evaluate_model(model, test_loader, device, class_names):
"""
Evaluates the model on a given dataloader (e.g., test set).
Computes confusion matrix and classification report.
"""
model.eval() # Set model to evaluation mode
all_preds = []
all_labels = []
with torch.no_grad(): # Disable gradient calculation
for images, labels in test_loader:
images, labels = images.to(device), labels.to(device)
outputs = model(images)
_, predictions = torch.max(outputs, 1)
all_preds.extend(predictions.cpu().numpy())
all_labels.extend(labels.cpu().numpy())
all_preds = np.array(all_preds)
all_labels = np.array(all_labels)
# Calculate confusion matrix
cm = confusion_matrix(all_labels, all_preds)
# Calculate classification report
class_report = classification_report(
all_labels, all_preds,
target_names=class_names,
digits=4,
zero_division=0
)
# Calculate overall accuracy from the report
accuracy = np.trace(cm) / np.sum(cm) # Simple accuracy from confusion matrix
return {
'confusion_matrix': cm,
'classification_report': class_report,
'accuracy': accuracy,
'predictions': all_preds,
'true_labels': all_labels
}
# Evaluate the model
eval_results = evaluate_model(model, test_loader, device, class_names)
# Print results
print("Classification Report:")
print(eval_results['classification_report'])
print(f"\nOverall Accuracy: {eval_results['accuracy']:.4f}")You should see output similar to:
Classification Report:
precision recall f1-score support
NORMAL 0.9780 0.3803 0.5477 234
PNEUMONIA 0.7280 0.9949 0.8407 390
accuracy 0.7644 624
macro avg 0.8530 0.6876 0.6942 624
weighted avg 0.8217 0.7644 0.7308 624
Overall Accuracy: 0.7644Visualizing the Confusion Matrix
A confusion matrix provides a clear visual representation of our model's performance:
def plot_confusion_matrix(confusion_matrix, class_names):
plt.figure(figsize=(8, 6))
sns.heatmap(
confusion_matrix,
annot=True,
fmt='d',
cmap='Blues',
xticklabels=class_names,
yticklabels=class_names
)
plt.xlabel('Predicted Label')
plt.ylabel('True Label')
plt.title('Confusion Matrix')
plt.tight_layout()
plt.show()
# Plot confusion matrix
plot_confusion_matrix(eval_results['confusion_matrix'], class_names)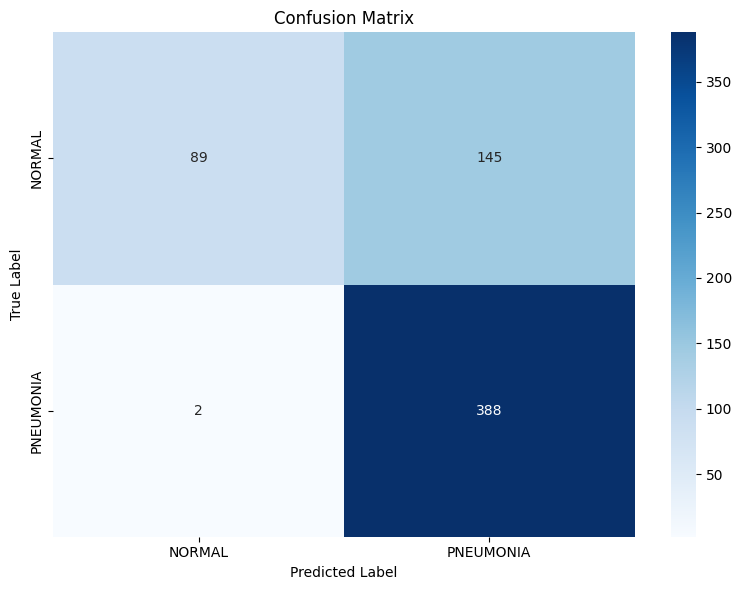
The confusion matrix shows:
- True Negatives (top-left): Normal X-rays correctly identified as normal
- False Positives (top-right): Normal X-rays incorrectly identified as pneumonia
- False Negatives (bottom-left): Pneumonia X-rays incorrectly identified as normal
- True Positives (bottom-right): Pneumonia X-rays correctly identified as pneumonia
Interpreting Results in a Medical Context
In medical diagnosis, different types of errors have different consequences:
- False Negatives (missing pneumonia): These are particularly dangerous as a patient with pneumonia might not receive necessary treatment, potentially leading to serious complications. Minimizing these is often a high priority (i.e., maximizing Recall/Sensitivity for the PNEUMONIA class).
- False Positives (diagnosing pneumonia when it's absent): These may lead to unnecessary treatment, causing stress and potential side effects, but are generally less immediately harmful than false negatives. Minimizing these relates to maximizing Recall/Specificity for the NORMAL class.
Examining our actual results from the Classification Report above, we see:
- Pneumonia Detection (Class 1): The model achieves extremely high Recall (Sensitivity) of ~0.9949. This is excellent, meaning it correctly identifies nearly 99.5% of the actual pneumonia cases in the test set, effectively minimizing dangerous False Negatives. However, its Precision for pneumonia is ~0.7280, meaning that when it predicts pneumonia, it's correct only about 73% of the time – the other 27% are False Positives (NORMAL cases misclassified as PNEUMONIA).
- Normal Case Detection (Class 0): The model still has very low Recall (Specificity) of ~0.3803. This indicates it only correctly identifies about 38% of the actual normal cases; the remaining 62% are misclassified as pneumonia (contributing to the lower precision for the PNEUMONIA class). The Precision for normal cases remains high (~0.9780), meaning if it predicts normal, it's very likely correct, but this model rarely makes that prediction for normal cases.
Interpretation: These results indicate the model is significantly biased towards predicting PNEUMONIA. It's highly sensitive but lacks specificity.
In a real medical scenario:
- The high sensitivity (~99.5%) is valuable for ensuring potential cases aren't missed.
- The low specificity (~38%) remains highly problematic, likely leading to a large number of unnecessary follow-ups for healthy individuals.
While prioritizing sensitivity is common for screening, this level of specificity would likely be impractical. These results strongly suggest that the class imbalance in our training data is heavily influencing the model's predictions. To create a more balanced and clinically useful model, addressing this imbalance directly (using techniques like weighted loss or resampling, as discussed in Section 5) would be the most logical next step.
4. Visualizing Model Predictions
Let's visualize some of our model's predictions to better understand its behavior:
def visualize_predictions(model, dataloader, device, class_names, num_samples=8):
"""Displays a batch of test images with their true labels and model predictions."""
model.eval()
try:
images, labels = next(iter(dataloader))
except StopIteration:
print("DataLoader is empty.")
return
# Ensure we don't request more samples than available in the batch
actual_samples = min(num_samples, images.size(0))
if actual_samples <= 0:
print("No samples in batch to display.")
return
images, labels = images[:actual_samples], labels[:actual_samples]
images_device = images.to(device) # Move input data to the correct device
# Get model predictions
with torch.no_grad():
outputs = model(images_device)
_, preds = torch.max(outputs, 1)
probs = F.softmax(outputs, dim=1)
# Move data back to CPU for plotting
preds = preds.cpu().numpy()
probs = probs.cpu().numpy()
images = images.cpu()
# Determine subplot layout
if actual_samples <= 4:
ncols = actual_samples; nrows = 1; figsize = (4 * ncols, 5)
else:
ncols = 4; nrows = 2; figsize = (16, 10)
fig, axes = plt.subplots(nrows=nrows, ncols=ncols, figsize=figsize)
# Ensure axes is iterable
if nrows == 1 and ncols == 1: axes = np.array([axes])
axes = axes.flatten()
fig.suptitle("Sample Test Set Predictions", fontsize=16)
for i, ax in enumerate(axes):
if i < actual_samples:
img_tensor = images[i]
true_label = class_names[labels[i]]
pred_label = class_names[preds[i]]
confidence = probs[i][preds[i]]
# Prepare image for display (C, H, W) -> (H, W, C)
img_display = img_tensor.permute(1, 2, 0).numpy()
# Display image
ax.imshow(img_display.squeeze(), cmap='gray', vmin=0.0, vmax=1.0)
# Set title with prediction info and color coding
title_color = 'green' if pred_label == true_label else 'red'
title = f"True: {true_label}\nPred: {pred_label}\nConf: {confidence:.2f}"
ax.set_title(title, color=title_color)
ax.axis('off')
else:
ax.axis('off')
plt.tight_layout()
plt.subplots_adjust(top=0.92) # Adjust layout for suptitle
plt.show()
# Visualize model predictions on the test set
print("\nVisualizing sample predictions from the test set...")
# Create a TEMPORARY DataLoader with shuffling enabled JUST for visualization
# This helps ensure we see a mix of classes in the first batch we grab.
# The 'test_loader' used for actual evaluation remains unshuffled.
temp_vis_loader = DataLoader(
dataset=test_dataset, # Use the same test_dataset
batch_size=batch_size, # Use the same batch size
shuffle=True # Shuffle ON for this temporary loader
)
visualize_predictions(model, temp_vis_loader, device, class_names)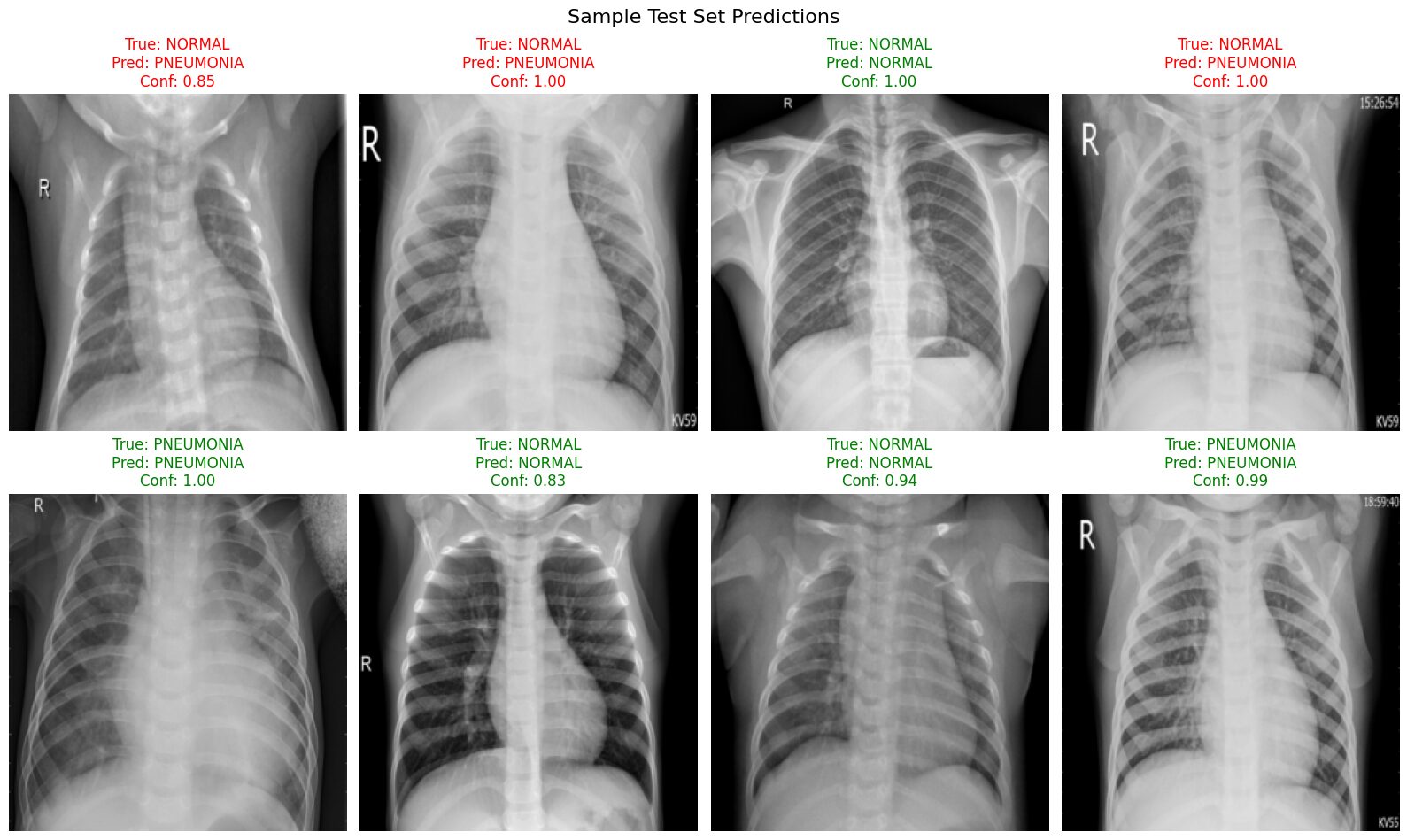
This visualization provides concrete examples of the model's behavior on the test set:
- We can clearly see examples of both correct (green titles) and incorrect (red titles) predictions made by the model.
- It allows us to observe the model's confidence for each prediction. Notice in this batch that the confidence scores are generally quite high (often >0.80), even for some of the incorrect classifications.
- Most importantly, we can identify potential patterns in the errors. In this specific sample batch, the errors primarily consist of
True: NORMALimages being incorrectly classified asPNEUMONIA, sometimes with high confidence. This visually reinforces the low Specificity (low Recall for the NORMAL class) identified in our quantitative evaluation metrics and highlights the model's tendency to misclassify normal cases.
Making Predictions on Random Images
Evaluating metrics like precision and recall gives us an overall sense of performance, but looking at individual predictions can provide more intuition. Let's see how our trained model performs on a randomly selected individual X-ray image from the NORMAL class in the test set.
First, here's the helper function we'll use to load, preprocess, and get a prediction for a single image path:
def predict_image(model, image_path, transform, device, class_names):
"""Loads a single image, preprocesses it, and returns model prediction details."""
try:
# Load the image using PIL
image = Image.open(image_path)
except FileNotFoundError:
print(f"Error: Image file not found at {image_path}")
return None
except Exception as e:
print(f"Error opening image {image_path}: {e}")
return None
# Preprocess: Apply validation/test transforms, add batch dimension, move to device
image_tensor = transform(image).unsqueeze(0).to(device)
# Make prediction
model.eval() # Ensure model is in evaluation mode
with torch.no_grad(): # Disable gradient calculations
output = model(image_tensor) # Output raw logits
probabilities = F.softmax(output, dim=1) # Probabilities
# Get the highest probability score and the corresponding class index
confidence, predicted_class_idx = torch.max(probabilities, 1)
# Extract results
class_idx = predicted_class_idx.item()
class_name = class_names[class_idx] # Map index to class name
confidence_score = confidence.item()
# Return results as a dictionary
return {
'class_id': class_idx,
'class_name': class_name,
'confidence': confidence_score,
'probabilities': probabilities[0].cpu().numpy() # All class probabilities
}Now, let's use this function on a random image from the test/NORMAL directory:
try:
normal_dir = os.path.join(test_dir, "NORMAL") # Target the NORMAL directory
# Get only .jpeg files from the directory
normal_test_files = [f for f in os.listdir(normal_dir) if f.lower().endswith('.jpeg')]
if not normal_test_files:
print(f"No NORMAL test images found in {normal_dir}.")
else:
# Select a random image file
random_filename = random.choice(normal_test_files)
test_image_path = os.path.join(normal_dir, random_filename)
print(f"\nPredicting on random NORMAL image: {random_filename}")
# Get prediction using the function
result = predict_image(model, test_image_path, val_test_transforms, device, class_names)
if result:
# Display the prediction details
print(f" Actual class: NORMAL") # State the true class
print(f" Predicted class: {result['class_name']}")
print(f" Confidence: {result['confidence']:.4f}")
print(f" Class probabilities: Normal={result['probabilities'][0]:.4f}, Pneumonia={result['probabilities'][1]:.4f}")
# Visualize the image with prediction
try:
img = Image.open(test_image_path)
plt.figure(figsize=(6, 6))
plt.imshow(img, cmap='gray')
# Include TRUE label in title for clarity
plt.title(f"True: NORMAL | Prediction: {result['class_name']} ({result['confidence']:.4f})")
plt.axis('off')
plt.show()
except Exception as e:
print(f"Error displaying image {test_image_path}: {e}")
except FileNotFoundError:
print(f"Error: Directory {normal_dir} not found.")
except Exception as e:
print(f"An error occurred during prediction example: {e}")Predicting on random NORMAL image: IM-0011-0001-0001.jpeg
Actual class: NORMAL
Predicted class: PNEUMONIA
Confidence: 0.7053
Class probabilities: Normal=0.2947, Pneumonia=0.7053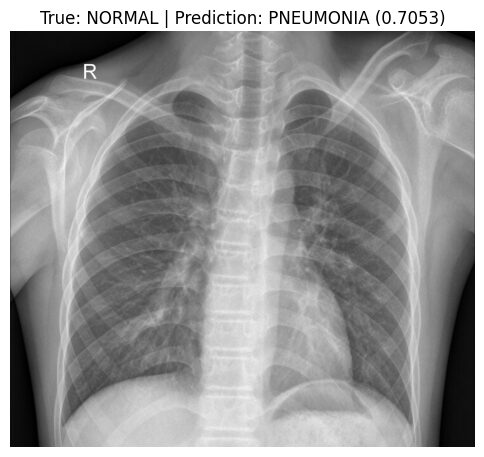
Here, we took a random test X-ray image (IM-0011-0001-0001.jpeg) known to be NORMAL. Our model, however, incorrectly predicted it as PNEUMONIA with moderate confidence (approx. 70.5%).
This specific misclassification provides a clear example of the main weakness identified in our evaluation metrics: the model's difficulty in correctly recognizing NORMAL cases (achieving only ~38.0% recall/specificity according to the Classification Report). Errors like this, where NORMAL images are falsely predicted as PNEUMONIA (False Positives), are why the overall Precision for the PNEUMONIA class was limited to ~72.8%. ****When the model predicts PNEUMONIA, roughly 27% of those predictions are actually NORMAL cases being misclassified.
While the model remains excellent at catching actual PNEUMONIA (with ~99.5% recall/sensitivity), this tendency to misclassify NORMAL images highlights the impact of the class imbalance. Looking at the specific image, we can see prominent normal structures (bronchial/vascular markings); it's plausible that the model, biased by the imbalance, struggles to differentiate these complex normal patterns from potential abnormalities.
Addressing this bias to improve specificity would clearly improve the model's clinical utility. This leads us nicely into exploring common training issues and techniques to mitigate them.
5. Addressing Common CNN Training Issues
Now that we've trained and evaluated our model, we've seen some promising results but also potential areas for improvement (like the low specificity driven by class imbalance). Let's explore common issues encountered during CNN development and strategies to address them, considering our specific pneumonia detection task.
Diagnosing and Addressing Overfitting
Overfitting occurs when a model learns the training data too well, including its noise and specific quirks, rather than the underlying general patterns. This leads to poor performance on new, unseen data. Signs of overfitting in our training plots would include:
- Training accuracy becoming much higher than validation accuracy.
- Training loss continuing to decrease significantly while validation loss plateaus or starts increasing.
Strategy: Early Stopping
If you observe validation loss starting to increase (a clear sign of overfitting), one effective strategy is early stopping.
- Concept: Monitor the validation loss after each epoch. Save the model's state whenever the validation loss reaches a new minimum. If the validation loss fails to improve for a predefined number of epochs (e.g., 5 or 10, known as "patience"), stop the training process. Finally, load the saved model state that achieved the best validation loss.
- Example Application: For our project, this would involve modifying the training loop to keep track of the best
epoch_val_lossseen so far, savingmodel.state_dict()at that point, and halting if the loss doesn't improve for the specified patience period.
Handling Underfitting
Underfitting is the opposite problem: the model fails to learn the training data well enough, resulting in poor performance on both the training and validation/test sets. This often suggests the model is too simple or hasn't trained sufficiently.
Potential Strategies:
- Increase Model Complexity: Make the model more powerful so it can capture more complex patterns.
- Example Application: We could add a fourth convolutional block to our
PneumoniaCNNdefinition or increase the number of output channels in the existingnn.Conv2dlayers (e.g., going from 32 -> 64 -> 128 to perhaps 64 -> 128 -> 256).
- Example Application: We could add a fourth convolutional block to our
- Train Longer: Give the model more time to learn by increasing the number of training epochs.
- Example Application: Simply call our
train_modelfunction with a larger value, likenum_epochs=30ornum_epochs=50, while carefully monitoring for signs of overfitting using the validation metrics.
- Example Application: Simply call our
- Reduce Regularization: Techniques like dropout prevent overfitting but can hinder learning if applied too aggressively when the model is underfitting.
- Example Application: We could try lowering the dropout probability in our fully connected layers, for instance, changing
nn.Dropout(p=0.5)tonn.Dropout(p=0.3). If using weight decay in the optimizer, we might reduce its strength.
- Example Application: We could try lowering the dropout probability in our fully connected layers, for instance, changing
- Learning Rate Adjustment: The learning rate might be too low (slow learning) or too high (preventing convergence). Experimenting or using a scheduler can help.
- Example Application: We could try initializing the
Adamoptimizer with a slightly different learning rate, likelr=0.001orlr=0.005. Alternatively, we could implement a learning rate scheduler (e.g.,torch.optim.lr_scheduler.ReduceLROnPlateau) that automatically reduces the learning rate if the validation loss stagnates.
- Example Application: We could try initializing the
Addressing Class Imbalance
As our verification step showed, the training set has a roughly 3:1 ratio of PNEUMONIA to NORMAL samples. This imbalance likely contributed to our model's bias towards predicting PNEUMONIA (high sensitivity, low specificity). Common strategies include:
- Weighted Loss Function: Modify the loss calculation to penalize errors on the minority class (NORMAL) more heavily.
- Example Application: Calculate weights inversely proportional to class frequency (e.g., assign a weight of ~3 to the NORMAL class and ~1 to the PNEUMONIA class) and pass these weights to the
weightparameter ofnn.CrossEntropyLosswhen defining ourcriterion.
- Example Application: Calculate weights inversely proportional to class frequency (e.g., assign a weight of ~3 to the NORMAL class and ~1 to the PNEUMONIA class) and pass these weights to the
- Resampling: Adjust the sampling process during training to create more balanced batches.
- Example Application: Oversampling the minority class involves drawing more samples (with replacement) from the NORMAL images during each epoch, perhaps using PyTorch's
WeightedRandomSamplerwith theDataLoader. Undersampling the majority class involves randomly discarding some PNEUMONIA samples to match the number of NORMAL samples, though this risks losing potentially useful information.
- Example Application: Oversampling the minority class involves drawing more samples (with replacement) from the NORMAL images during each epoch, perhaps using PyTorch's
- Generate Synthetic Data: Create artificial examples of the minority class.
- Example Application: This often involves more advanced techniques like SMOTE (Synthetic Minority Over-sampling Technique) or using Generative Adversarial Networks (GANs) to create new, realistic-looking NORMAL X-ray images, though implementing these is beyond the scope of this tutorial.
Choosing the right strategy often involves experimentation. For class imbalance, using a weighted loss or resampling via WeightedRandomSampler are often effective starting points.
Review and Next Steps
Congratulations! You've successfully navigated this two-part tutorial series, journeying from the fundamentals of Convolutional Neural Networks all the way to building, training, and evaluating a practical pneumonia detection model using PyTorch. You've seen the entire workflow, from defining the architecture in Part 1 to preparing data, implementing training loops, interpreting results, and considering common challenges in Part 2.
What You've Learned
As we wrap up, let's distill the most important concepts and practices covered across both tutorials:
- End-to-End Workflow: Building an effective computer vision solution involves a complete pipeline: careful data preparation (verification, splitting, augmentation, transformation), thoughtful model architecture definition (often using PyTorch's object-oriented
nn.Modulestructure), implementing robust training loops (managing device placement and model modes), and performing rigorous evaluation tailored to the problem. - Data is Foundational: The quality and handling of your data are paramount. Accurate verification, appropriate splitting (train/validation/test), deliberate preprocessing choices (like resizing or grayscale conversion), and techniques like data augmentation significantly impact model performance and reliability.
- Evaluate Beyond Accuracy: Especially for real-world applications like medical diagnosis, relying solely on accuracy can be misleading, particularly with imbalanced datasets. Metrics like precision, recall (sensitivity/specificity), F1-score, and confusion matrices provide a much deeper understanding of model strengths and weaknesses for each class.
- Practical Training Details Matter: Correctly switching between
model.train()andmodel.eval()is essential for layers like Dropout and BatchNorm to function properly. Being aware of potential issues like overfitting or class imbalance and knowing strategies to address them (e.g., early stopping, learning rate scheduling, weighted loss, resampling) are key practical skills for refining models.
What You Can Try Next
Your journey into computer vision with PyTorch doesn't have to end here! To deepen your skills, consider exploring these areas:
- Transfer Learning: Instead of training from scratch, leverage powerful models (like ResNet, VGG, DenseNet) pre-trained on large datasets (like ImageNet) and fine-tune them for your specific task. This often leads to better performance with less data and faster training.
- Cross-Validation: Implement k-fold cross-validation for a more robust evaluation of your model's performance, reducing the dependency on a single train-validation split.
- Hyperparameter Tuning: Systematically experiment with different learning rates, batch sizes, optimizer choices, network architectures, or augmentation strategies.
- Explainability: Use techniques like Grad-CAM, SHAP, or LIME to understand why your model makes certain predictions. Visualize the image regions that most influence its decision. This is important for building trust, especially in medical AI.
Remember that deep learning is as much an art as it is a science—experimentation, careful analysis, and domain knowledge all play important roles in creating effective solutions. Keep practicing these skills, and you'll be well-equipped to solve real-world problems with computer vision in PyTorch.
Additional Resources
To continue your learning journey:
- PyTorch Documentation - Comprehensive reference for all PyTorch functions
- Sequence Models in PyTorch - If you're interested in extending your skills to sequential data
- Natural Language Processing with PyTorch - For processing text data with PyTorch
- Medical Image Analysis Papers - Recent research in medical image classification